| << Chapter < Page | Chapter >> Page > |
We are attempting to classify and count the different subspecies of WBC in multi-cell images using morphological features. This is a challenging and ongoing problem explored by many independent researchers, and there is no standard solution. While shape and feature recognition is easy to humans, it is one of the most researched topics in machine learning. Most methods divide the process into two subsystems, segmentation and classification.
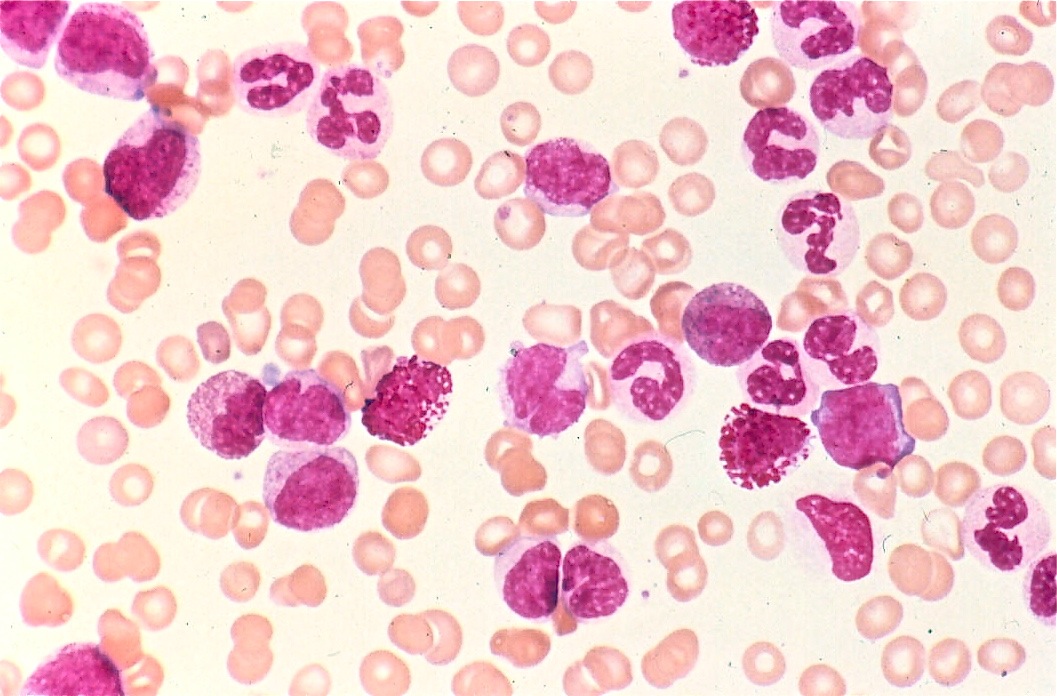
Figure 1. Overlapping can be seen between RBCs and WBCs.
Green channel filtering is a common way to isolate WBCs in colored blood smear images. Red blood cells (RBCs) and white blood cells separate into two distinct peaks in this space (with number of pixels on the y axis and color value, 0 to 255 on the x axis). The white blood cells have more green content. Furthermore, the WBC nuclei also differ slightly from the cytoplasm in green content. [1] However, this method requires extremely consistent and high quality images due to the subtleties associated with the different cells’ green channel content.
Grayscale images and thresholding can also be used to separate the RBCs, WBCs, and WBC nuclei. WBC cell bodies appear much darker than RBCs, and WBC nuclei appear darker than cell bodies. Several thresholds can be used to differentiate between the cells. [2] For example, in “White Blood Cell Segmentation Using Morphological Operators and Scale-Space Analysis,” a hard-coded threshold was used to isolate the first seed. Then, another equation was used to improve the gradient extracted. This was repeated for 10 iterations. [3]This method requires the images to be high resolution, because blur and graininess can cause individual areas of darker or lighter spots.
Some researchers use a combination of both common methods in order to obtain the best results. They first localize the nuclei using color analysis, and then threshold for finer boundaries. [1]
We found that many research articles used both morphological and texture features for classification. These are based mostly on nucleus shape. Recall that the subtypes of WBCs have lobed nuclei which differ between the types. In the literature, monocytes are the most difficult to classify, often mistaken lymphocytes and neutrophils.
Most research papers implement at least ten features. These features must differ significantly between the cell types. They include a variety of area, perimeter, and color characteristics. The papers that used colored images have more features than grayscale due to the differences in color content as mentioned above. Below are example categories that have been explored.
Common classification methods include k-nearest neighbors, learning vector quantization, Multi-layer perceptrons, and support vector machine (SVM). These are pattern recognition algorithms, which makes sense for this application. We found that the most successful paper, which used 57 features in total from all three categories above, had an accuracy of 91% for SVM. [3]

Notification Switch
Would you like to follow the 'Automatic white blood cell classification using svm and neural networks' conversation and receive update notifications?