| << Chapter < Page | Chapter >> Page > |
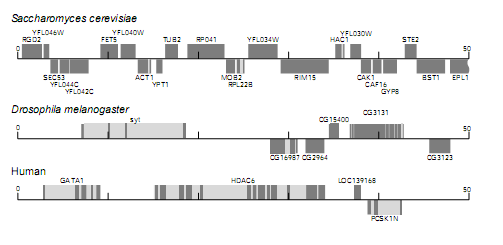
Identifying genes in DNA sequences from higher organisms is usually more difficult than in bacteria. This is because in humans, for example, gene coding sequences are separated by long sequences that do not code for proteins. Moreover, genes of higher eukaryotes intronsintrons are interrupted by introns, which are sequences that are spliced out of the NA before intronsintrons translation. The presence of introns breaks up the open reading frames into short segments, making them much harder to distinguish from non-coding sequences. The maps below show 50 kbp segments of DNA from yeast, Drosophila, and humans. The dark grey boxes represent coding sequences and the light grey boxes represent introns. The boxes above the line are transcribed to the right and the boxes below are transcribed to the left. Names have been assigned to each of the identified genes. Although the yeast genes are much like those of bacteria (few introns and packed closely together), the Drosophila and human genes are spread apart and interrupted by many introns. Sophisticated computer algorithms were used to identify these dispersed gene sequences.
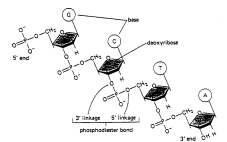
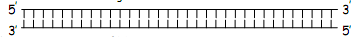
In a double stranded DNA molecule the two strands run anti-parallel to one another and the general structure can be diagramed like this:
Note about representation of DNA sequences:
1) Single strands are always represented in direction of synthesis 5’ to 3’.
2) For double stranded DNA, usually one strand is represented in the 5’ to 3’ direction.
For a gene, the strand represented would correspond to the sequence of the mRNA.
DNA polymersaes are the key players in the methods that we will be considering. The general reaction carried out by DNA polymerase is to synthesize a copy of a DNA template, starting with the chemical precursors (nucleotides) dATP, dGTP, dCTP, and dTTP (dNTPs).
All DNA polymerases have two fundamental properties in common:
(1) New DNA is synthesized only by elongation of an existing strand at its 3 end.
(2) Synthesis requires nucleotide precursors, a free 3 OH end, and a template strand.
A general substrate for DNA polymerase looks like this:

Note that the template strand can be as short as 1 base or as long as several thousand bases. After addition of DNA polymerase and nucleotide precursors, this product will be readily synthesized:

(1) The two DNA strands are separated. Heating to 100C to melt the base pairing hydrogen bonds that hold the strands together does this.
(2) A short oligonucleotide (ca. 18 bases) designed to be complimentary to the end of one of the strands is allowed to anneal to the single stranded DNA. The resulting DNA hybrid looks much like the general polymerase substrate shown previously.

Notification Switch
Would you like to follow the 'Genetics' conversation and receive update notifications?