| << Chapter < Page | Chapter >> Page > |
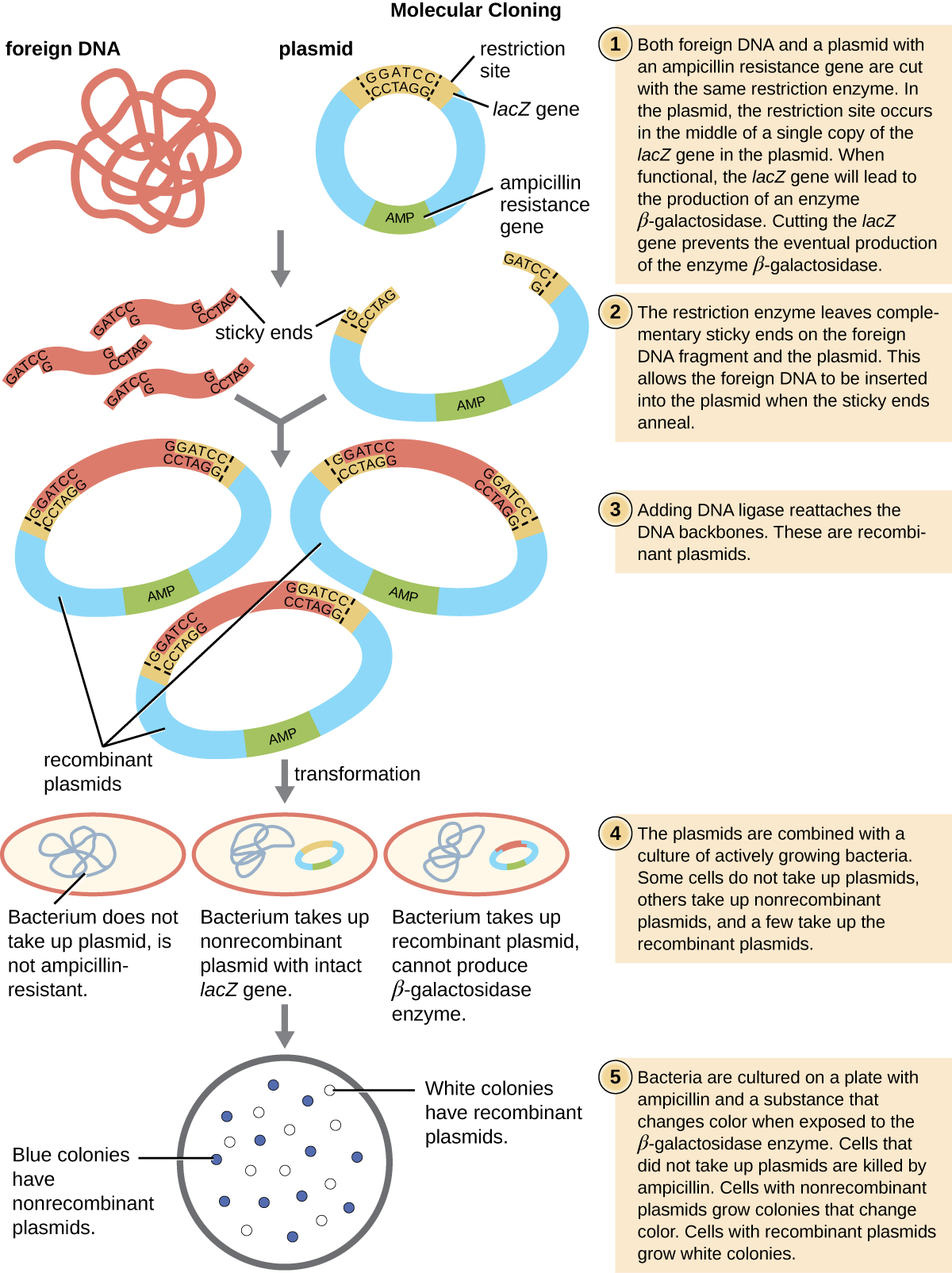
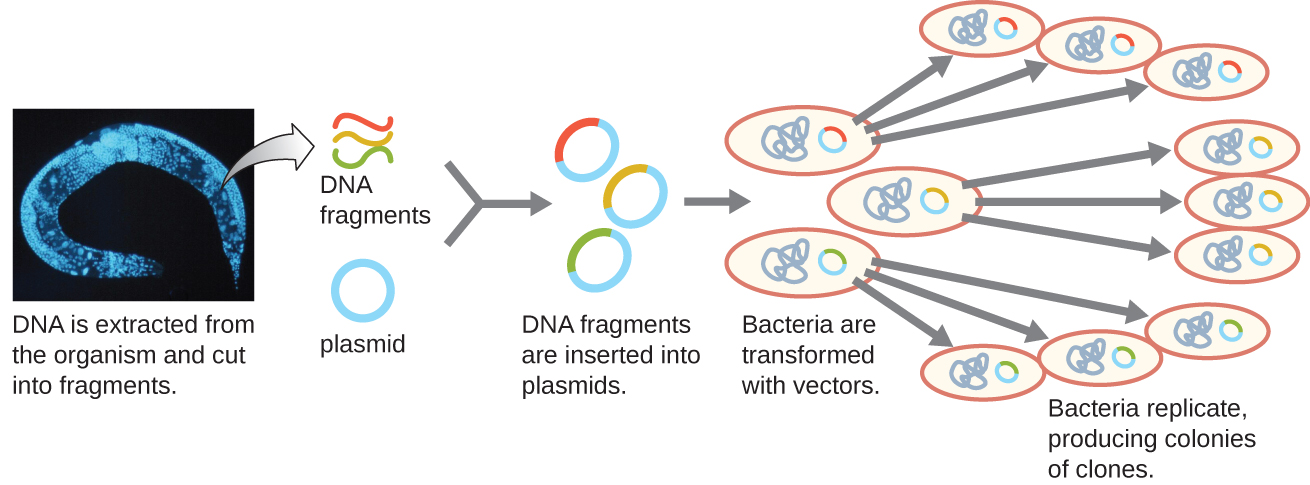
Molecular cloning may also be used to generate a genomic library . The library is a complete (or nearly complete) copy of an organism’s genome contained as recombinant DNA plasmids engineered into unique clones of bacteria. Having such a library allows a researcher to create large quantities of each fragment by growing the bacterial host for that fragment. These fragments can be used to determine the sequence of the DNA and the function of any genes present.
One method for generating a genomic library is to ligate individual restriction enzyme-digested genomic fragments into plasmid vectors cut with the same restriction enzyme ( [link] ). After transformation into a bacterial host, each transformed bacterial cell takes up a single recombinant plasmid and grows into a colony of cells. All of the cells in this colony are identical clones and carry the same recombinant plasmid. The resulting library is a collection of colonies, each of which contains a fragment of the original organism’s genome, that are each separate and distinct and can each be used for further study. This makes it possible for researchers to screen these different clones to discover the one containing a gene of interest from the original organism’s genome.
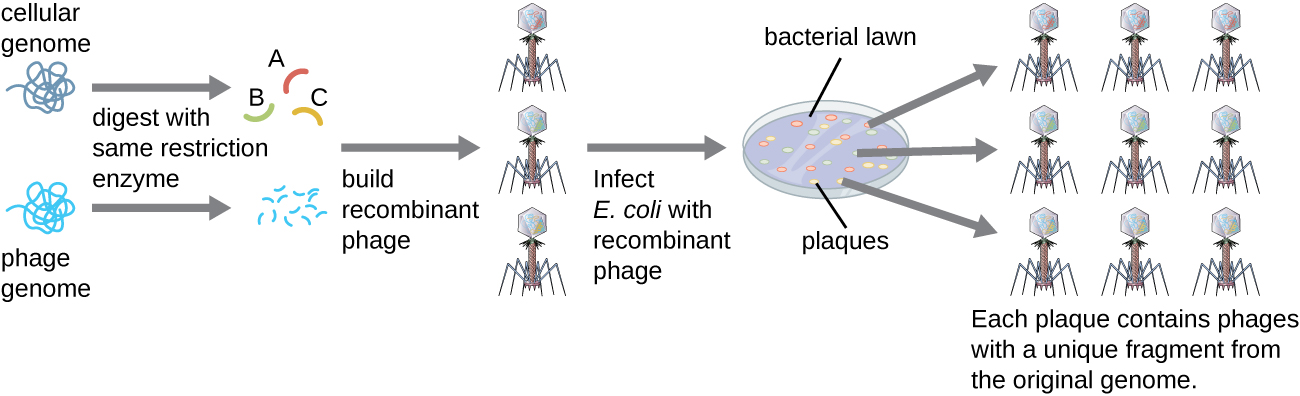
To construct a genomic library using larger fragments of genomic DNA, an E. coli bacteriophage, such as lambda , can be used as a host ( [link] ). Genomic DNA can be sheared or enzymatically digested and ligated into a pre-digested bacteriophage lambda DNA vector. Then, these recombinant phage DNA molecules can be packaged into phage particles and used to infect E. coli host cells on a plate. During infection within each cell, each recombinant phage will make many copies of itself and lyse the E. coli lawn, forming a plaque. Thus, each plaque from a phage library represents a unique recombinant phage containing a distinct genomic DNA fragment. Plaques can then be screened further to look for genes of interest. One advantage to producing a library using phages instead of plasmids is that a phage particle holds a much larger insert of foreign DNA compared with a plasmid vector, thus requiring a much smaller number of cultures to fully represent the entire genome of the original organism.

Notification Switch
Would you like to follow the 'Microbiology' conversation and receive update notifications?