| << Chapter < Page | Chapter >> Page > |
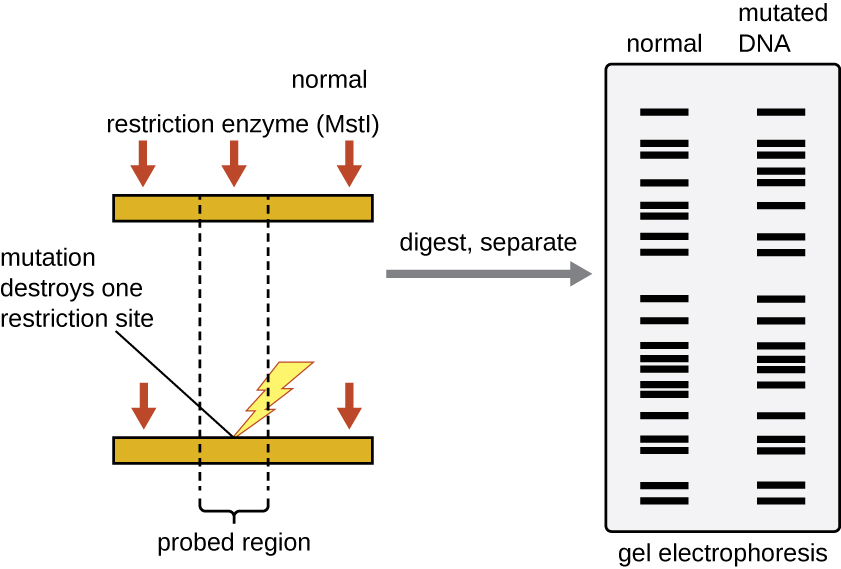
Forensic scientists use RFLP analysis as a form of DNA fingerprinting , which is useful for analyzing DNA obtained from crime scenes, suspects, and victims. DNA samples are collected, the numbers of copies of the sample DNA molecules are increased using PCR , and then subjected to restriction enzyme digestion and agarose gel electrophoresis to generate specific banding patterns. By comparing the banding patterns of samples collected from the crime scene against those collected from suspects or victims, investigators can definitively determine whether DNA evidence collected at the scene was left behind by suspects or victims.
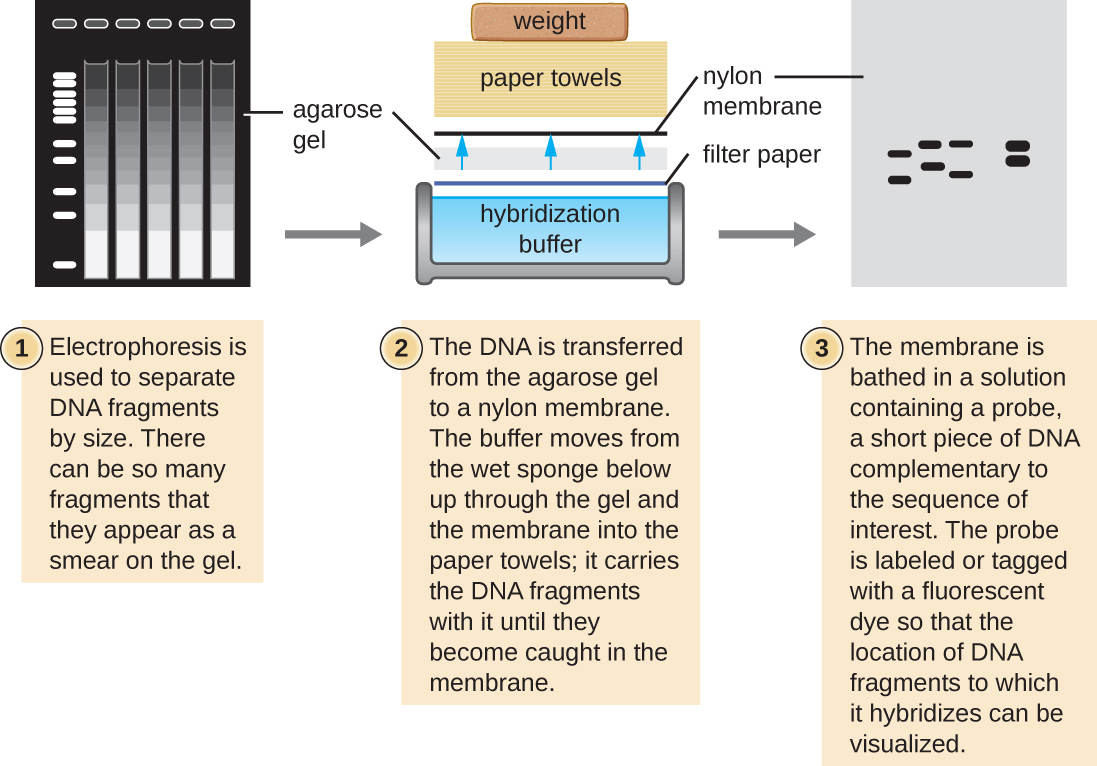
Several molecular techniques capitalize on sequence complementarity and hybridization between nucleic acids of a sample and DNA probes. Typically, probing nucleic-acid samples within a gel is unsuccessful because as the DNA probe soaks into a gel, the sample nucleic acids within the gel diffuse out. Thus, blotting techniques are commonly used to transfer nucleic acids to a thin, positively charged membrane made of nitrocellulose or nylon. In the Southern blot technique, developed by Sir Edwin Southern in 1975, DNA fragments within a sample are first separated by agarose gel electrophoresis and then transferred to a membrane through capillary action ( [link] ). The DNA fragments that bind to the surface of the membrane are then exposed to a specific single-stranded DNA probe labeled with a radioactive or fluorescent molecular beacon to aid in detection. Southern blots may be used to detect the presence of certain DNA sequences in a given DNA sample. Once the target DNA within the membrane is visualized, researchers can cut out the portion of the membrane containing the fragment to recover the DNA fragment of interest.
Variations of the Southern blot—the dot blot, slot blot, and the spot blot—do not involve electrophoresis, but instead concentrate DNA from a sample into a small location on a membrane. After hybridization with a DNA probe, the signal intensity detected is measured, allowing the researcher to estimate the amount of target DNA present within the sample.
A colony blot is another variation of the Southern blot in which colonies representing different clones in a genomic library are transferred to a membrane by pressing the membrane onto the culture plate. The cells on the membrane are lysed and the membrane can then be probed to determine which colonies within a genomic library harbor the target gene. Because the colonies on the plate are still growing, the cells of interest can be isolated from the plate.

Notification Switch
Would you like to follow the 'Microbiology' conversation and receive update notifications?