| << Chapter < Page | Chapter >> Page > |
The prokaryotes, which include bacteria and archaea, are mostly single-celled organisms that, by definition, lack membrane-bound nuclei and other organelles. A bacterial chromosome is a covalently closed circle that, unlike eukaryotic chromosomes, is not organized around histone proteins. The central region of the cell in which prokaryotic DNA resides is called the nucleoid. In addition, prokaryotes often have abundant plasmids , which are shorter circular DNA molecules that may only contain one or a few genes. Plasmids can be transferred independently of the bacterial chromosome during cell division and often carry traits such as antibiotic resistance.
Transcription in prokaryotes (and in eukaryotes) requires the DNA double helix to partially unwind in the region of mRNA synthesis. The region of unwinding is called a transcription bubble. Transcription always proceeds from the same DNA strand for each gene, which is called the template strand . The mRNA product is complementary to the template strand and is almost identical to the other DNA strand, called the nontemplate strand . The only difference is that in mRNA, all of the T nucleotides are replaced with U nucleotides. In an RNA double helix, A can bind U via two hydrogen bonds, just as in A–T pairing in a DNA double helix.
The nucleotide pair in the DNA double helix that corresponds to the site from which the first 5' mRNA nucleotide is transcribed is called the +1 site, or the initiation site . Nucleotides preceding the initiation site are given negative numbers and are designated upstream . Conversely, nucleotides following the initiation site are denoted with “+” numbering and are called downstream nucleotides.
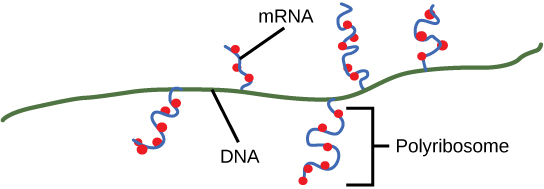
Prokaryotes do not have membrane-enclosed nuclei. Therefore, the processes of transcription, translation, and mRNA degradation can all occur simultaneously.
Our discussion here will exemplify transcription by describing this process in Escherichia coli , a well-studied bacterial species. Although some differences exist between transcription in E. coli and transcription in archaea, an understanding of E. coli transcription can be applied to virtually all bacterial species.
A promoter is a DNA sequence onto which the transcription machinery binds and initiates transcription. In most cases, promoters exist upstream of the genes they regulate. The specific sequence of a promoter is very important because it determines whether the corresponding gene is transcribed all the time, some of the time, or infrequently. Although promoters vary among prokaryotic genomes, a few elements are conserved.
View this MolecularMovies animation to see the first part of transcription and the base sequence repetition of the TATA box.
The transcription elongation phase begins with RNA polymerase synthesizing mRNA in the 5' to 3' direction at a rate of approximately 40 nucleotides per second. As elongation proceeds, the DNA is continuously unwound ahead of the core enzyme and rewound behind it ( [link] ). The base pairing between DNA and RNA is not stable enough to maintain the stability of the mRNA synthesis components. Instead, the RNA polymerase acts as a stable linker between the DNA template and the nascent RNA strands to ensure that elongation is not interrupted prematurely.
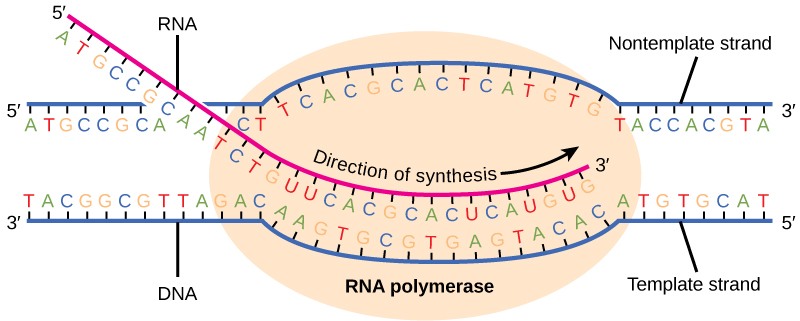
Once a gene is transcribed, the prokaryotic polymerase needs to be instructed to dissociate from the DNA template and liberate the newly made mRNA. Depending on the gene being transcribed, there are two kinds of termination signals. One is protein-based and the other is RNA-based. Rho-dependent termination is controlled by the rho protein, which tracks along behind the polymerase on the growing mRNA chain. Near the end of the gene, the polymerase encounters a run of G nucleotides on the DNA template and it stalls. As a result, the rho protein collides with the polymerase. The interaction with rho releases the mRNA from the transcription bubble.
Rho-independent termination is controlled by specific sequences in the DNA template strand. As the polymerase nears the end of the gene being transcribed, it encounters a region rich in C–G nucleotides. The mRNA folds back on itself, and the complementary C–G nucleotides bind together. The result is a stable hairpin that causes the polymerase to stall as soon as it begins to transcribe a region rich in A–T nucleotides. The complementary U–A region of the mRNA transcript forms only a weak interaction with the template DNA. This, coupled with the stalled polymerase, induces enough instability for the core enzyme to break away and liberate the new mRNA transcript.
Upon termination, the process of transcription is complete. By the time termination occurs, the prokaryotic transcript would already have been used to begin synthesis of numerous copies of the encoded protein because these processes can occur concurrently. The unification of transcription, translation, and even mRNA degradation is possible because all of these processes occur in the same 5' to 3' direction, and because there is no membranous compartmentalization in the prokaryotic cell ( [link] ). In contrast, the presence of a nucleus in eukaryotic cells prevents simultaneous transcription and translation.
Visit this BioStudio animation to see the process of prokaryotic transcription.
In prokaryotes, mRNA synthesis is initiated at a promoter sequence on the DNA template comprising two consensus sequences that recruit RNA polymerase. The prokaryotic polymerase consists of a core enzyme of four protein subunits and a σ protein that assists only with initiation. Elongation synthesizes mRNA in the 5' to 3' direction at a rate of 40 nucleotides per second. Termination liberates the mRNA and occurs either by rho protein interaction or by the formation of an mRNA hairpin.

Notification Switch
Would you like to follow the 'General biology part i - mixed majors' conversation and receive update notifications?