| << Chapter < Page | Chapter >> Page > |
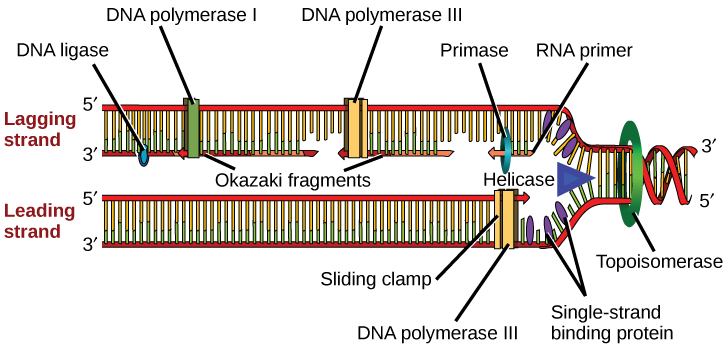
You isolate a cell strain in which the joining together of Okazaki fragments is impaired and suspect that a mutation has occurred in an enzyme found at the replication fork. Which enzyme is most likely to be mutated?
The replication fork moves at the rate of 1000 nucleotides per second. DNA polymerase can only extend in the 5' to 3' direction, which poses a slight problem at the replication fork. As we know, the DNA double helix is anti-parallel; that is, one strand is in the 5' to 3' direction and the other is oriented in the 3' to 5' direction. One strand, which is complementary to the 3' to 5' parental DNA strand, is synthesized continuously towards the replication fork because the polymerase can add nucleotides in this direction. This continuously synthesized strand is known as the leading strand . The other strand, complementary to the 5' to 3' parental DNA, is extended away from the replication fork, in small fragments known as Okazaki fragments , each requiring a primer to start the synthesis. Okazaki fragments are named after the Japanese scientist who first discovered them. The strand with the Okazaki fragments is known as the lagging strand .
The leading strand can be extended by one primer alone, whereas the lagging strand needs a new primer for each of the short Okazaki fragments. The overall direction of the lagging strand will be 3' to 5', and that of the leading strand 5' to 3'. A protein called the sliding clamp holds the DNA polymerase in place as it continues to add nucleotides. The sliding clamp is a ring-shaped protein that binds to the DNA and holds the polymerase in place. Topoisomerase prevents the over-winding of the DNA double helix ahead of the replication fork as the DNA is opening up; it does so by causing temporary nicks in the DNA helix and then resealing it. As synthesis proceeds, the RNA primers are replaced by DNA. The primers are removed by the exonuclease activity of DNA pol I, and the gaps are filled in by deoxyribonucleotides. The nicks that remain between the newly synthesized DNA (that replaced the RNA primer) and the previously synthesized DNA are sealed by the enzyme DNA ligase that catalyzes the formation of phosphodiester linkage between the 3'-OH end of one nucleotide and the 5' phosphate end of the other fragment.
Once the chromosome has been completely replicated, the two DNA copies move into two different cells during cell division. The process of DNA replication can be summarized as follows:
[link] summarizes the enzymes involved in prokaryotic DNA replication and the functions of each.
| Prokaryotic DNA Replication: Enzymes and Their Function | |
|---|---|
| Enzyme/protein | Specific Function |
| DNA pol I | Exonuclease activity removes RNA primer and replaces with newly synthesized DNA |
| DNA pol II | Repair function |
| DNA pol III | Main enzyme that adds nucleotides in the 5'-3' direction |
| Helicase | Opens the DNA helix by breaking hydrogen bonds between the nitrogenous bases |
| Ligase | Seals the gaps between the Okazaki fragments to create one continuous DNA strand |
| Primase | Synthesizes RNA primers needed to start replication |
| Sliding Clamp | Helps to hold the DNA polymerase in place when nucleotides are being added |
| Topoisomerase | Helps relieve the stress on DNA when unwinding by causing breaks and then resealing the DNA |
| Single-strand binding proteins (SSB) | Binds to single-stranded DNA to avoid DNA rewinding back. |
Review the full process of DNA replication here .
Replication in prokaryotes starts from a sequence found on the chromosome called the origin of replication—the point at which the DNA opens up. Helicase opens up the DNA double helix, resulting in the formation of the replication fork. Single-strand binding proteins bind to the single-stranded DNA near the replication fork to keep the fork open. Primase synthesizes an RNA primer to initiate synthesis by DNA polymerase, which can add nucleotides only in the 5' to 3' direction. One strand is synthesized continuously in the direction of the replication fork; this is called the leading strand. The other strand is synthesized in a direction away from the replication fork, in short stretches of DNA known as Okazaki fragments. This strand is known as the lagging strand. Once replication is completed, the RNA primers are replaced by DNA nucleotides and the DNA is sealed with DNA ligase, which creates phosphodiester bonds between the 3'-OH of one end and the 5' phosphate of the other strand.
[link] You isolate a cell strain in which the joining together of Okazaki fragments is impaired and suspect that a mutation has occurred in an enzyme found at the replication fork. Which enzyme is most likely to be mutated?
[link] DNA ligase, as this enzyme joins together Okazaki fragments.

Notification Switch
Would you like to follow the 'Biology' conversation and receive update notifications?